Our lab focuses on understanding mechanisms of resistance to therapies for Gastrointestinal malignancies. We have access to thousands of patient samples from which we can investigate driving mutations in the tumor, understand changes in RNA expression with and without therapy, and understand and characterize the tumor microenvironment. We work closely with, and have a need, for computational biology. We validate any observations in preclinical models, and develop novel co-culture models to test cell-cell interactions. The aim is to refine our understanding of disease biology to effect therapeutic opportunities for patients with cancer.
Recent projects have included:
(1) Understanding the mechanism of taxane resistance in upper gastrointestinal malignancies (in collaboration with the Giannakakou Lab).
This collaboration led to the discovery of CLIP-170s – a short isoform of the plus-tip microtubule trafficking protein – that altered the availability of taxane binding to microtubules and leads to relative resistance to taxane therapy. The collaboration included real-time confocal live imaging, computational analyses, knock-in and knock-out experimentation, and correlation with clinical samples. This highly translational project is ongoing and will hopefully lead to a novel therapeutic strategy. This was NIH R01 supported.
(2) Evaluating the gastric microbiome and implications for malignancy.
We developed novel methodology to examine by WGS the microbial content of endoscopic tissue biopsies in patients with and without gastric cancer. We discovered the diversity of the microbiome in gastric cancer, and how it is associated with cancer progression. This research was in close collaboration with computational biology, where we were the first to evaluate an ability to identify and characterize the microbiome from small endoscopic biopsy samples (ref). This was funded by the STARR cancer foundation.
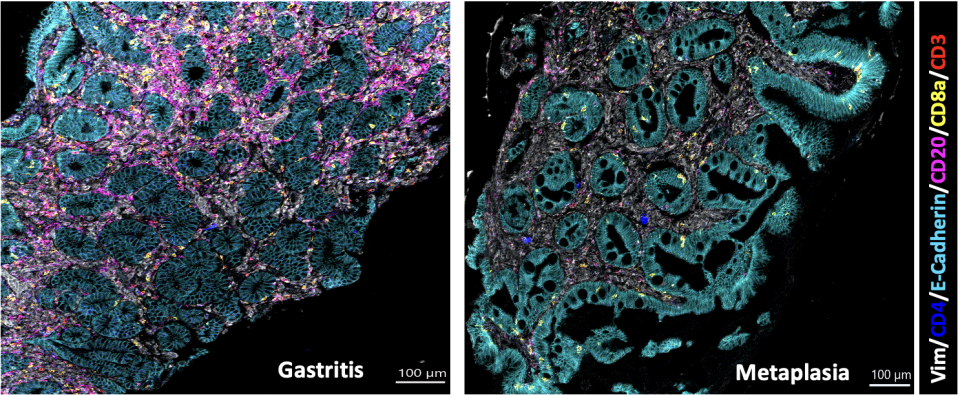
(3) Characterizing the tumor microenvironment of gastric cancer.
Gastric cancer has been well established to have several disease subtypes – historically, we know of the Laurens classification of Diffuse, Intestinal, and mixed categories. More recently, with next generation sequencing, we have identified an microsatellite-high, genomically stable, chromosomal instable, and Epstein-Barr virus subtype (TCGA classification). Partnering with Boston Gene, we have characterized the tumor microenvironment of gastric cancer into 5 distinct subtypes – immune enriched, B-cell inflamed, immune depleted, fibrotic, and mesenchymal. This discovery has important clinical implications for immunotherapy. This project and line of research was funded by philanthropy.
Gastric Microenvironment
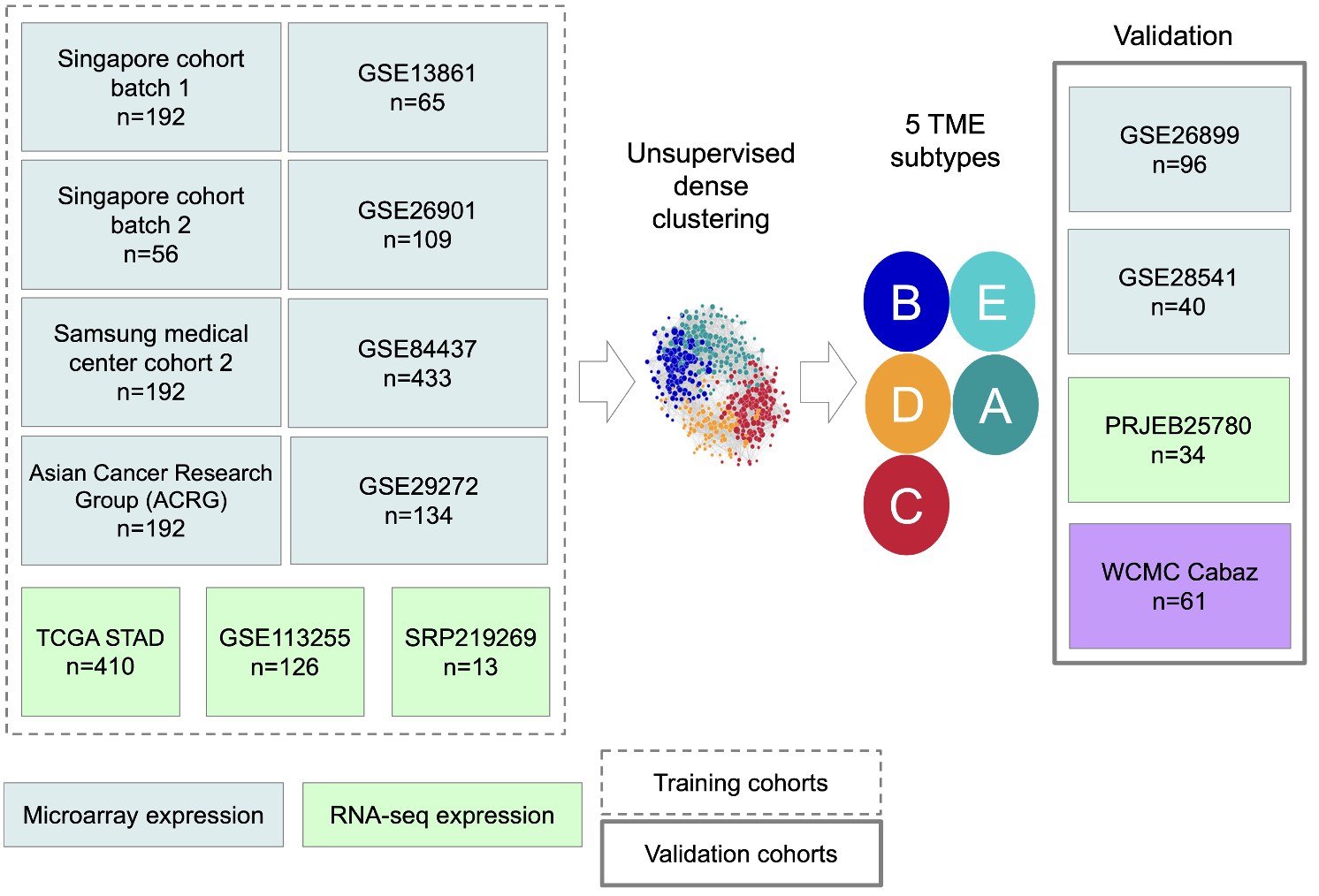
In collaboration with Boston Gene Corp, the Shah Lab is attempting to classify gastric tumor microenvironments in a way that is more clinically relevant.
Taxane Resistance
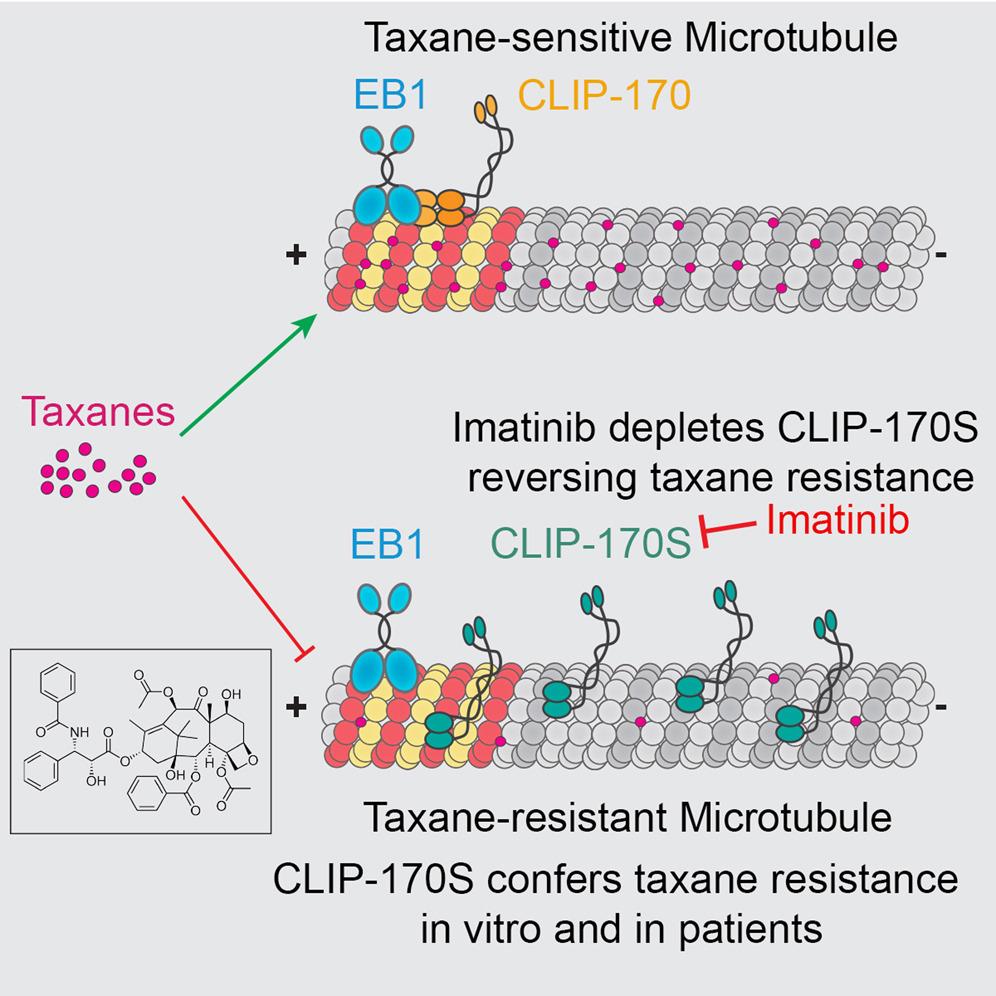
Working with the Giannakakou Lab, we developed a model showing CLIP-170S as a microtubule+TIP variant conferring taxane resistance. Read more below: